
Wang Z, Zhao F, Peng J, Xu J (2011) Protein 8-class secondary structure prediction using conditional neural fields. Proteins Struct Funct Bioinform 47(2):228–235ĭrozdetskiy A, Cole C, Procter J, Barton GJ (2015) JPred4: a protein secondary structure prediction server. Pollastri G, Przybylski D, Rost B, Baldi P (2002) Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. Proteins Struct Funct Bioinform 19(1):55–72 Rost B, Sander C (1994) Combining evolutionary information and neural networks to predict protein secondary structure. Garnier J, Gibrat JF, Robson B (1996) GOR method for predicting protein secondary structure from amino acid sequence.
Geourjon C, Deleage G (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Doctoral dissertation, University of PittsburghĬhou PY, Fasman GD (1974) Prediction of protein conformation. Hoye AT (2010) Synthesis of natural and non-natural polycylicalkaloids. The proposed model shows an average improvement of ~ 2% and ~ 3% accuracy over the existing methods in a blind test for Q8 and Q3 accuracy. The ensembling technique used along with variability in datasets can remove the bias of each dataset by balancing it and making the features more distinguishable, leading to the improvement in accuracy as compared to the conventional and existing techniques.

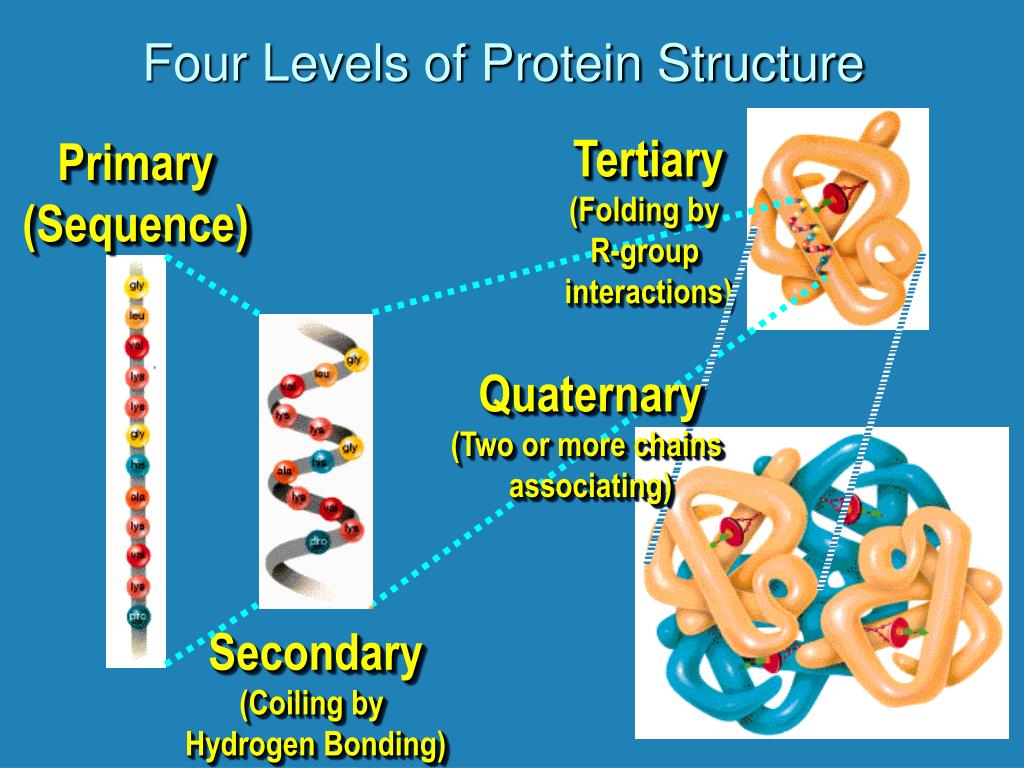
The model is evaluated using performance measures: Q8 and Q3 cross-validation accuracy, class precision, class recall, kappa factor, and testing on a dataset that is not used for training purposes, i.e., blind test. These standard datasets possess less than 25% redundancy. The data from standard datasets, namely CB513, RS126, PTOP742, PSA472, and MANESH, have been used for training and testing purposes. In the present study, ensemble techniques such as AdaBoost- and Bagging-based deep learning models are proposed to predict the protein secondary structure. Thus, there is an excellent room for developing new models of protein secondary structure prediction to address the issues of prediction accuracy. There exists a vast literature on the protein secondary structure prediction approaches (more than 20 techniques), but to date, none of the existing techniques is entirely accurate. Assigning the local conformation to protein sequences requires much computational work. \) in the side chain of hydroxyproline allows more hydrogen bonding in the collagen peptide which makes connective tissues stronger.Protein secondary structure is the local conformation assigned to protein sequences with the help of its three-dimensional structure.


 0 kommentar(er)
0 kommentar(er)
